Parameter Optimization of an Ecological Niche Modeling Workflow
Optimization Research Object Summary and Description
The Ecological Niche Modeling (ENM) workflow has been designed to perform the analysis of species distributions and to predict changes in biodiversity patterns [Kulhanek et al 2011, Wiley et al 2003, Guinan et al 2009]. The idea of niche modeling is based on G.E. Hutchinson's definition of the realized niche, where a set of environmental factors or a multidimensional space of resources (e.g. light and structure), can be used to predict the persistence of a species [Hutchinson 1957]. Thus, potential distribution models can be generated with relatively few variables characterizing the abiotic environment of the species in the form of geo-referenced raster layers.
In order to obtain valid models for species niches, in many cases it is important to specify the appropriate set of parameter values for a given input data set (occurrences, mask, and layers). The best parameter values can vary between different input data sets and hence are difficult to know beforehand.
The workflow uses web services to remotely execute a specific modeling algorithm, provided by the openModeller tool. The result model represents the suitable conditions of abiotic for a given species. After creating the model, a test model operation is called. This operation tests the model, by using the 10% points left out of model creation. The test operation calculates the receiver operating characteristic (ROC) curve, area under the curve (AUC). An abstract representation of the workflow can be seen below:
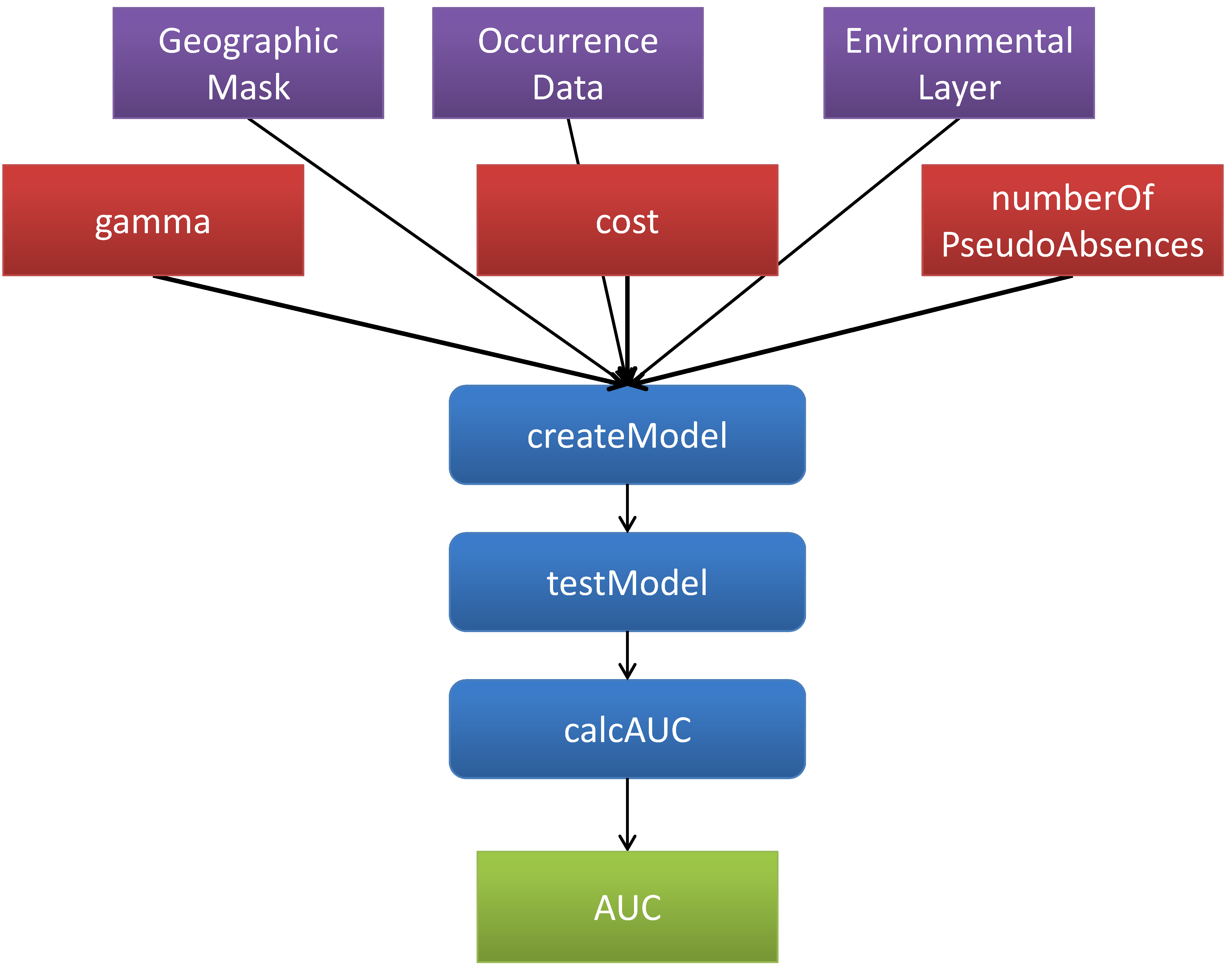
This page represents a Research Object representing the optimizations made on the AUC output parameter of the ENM workflow using Support Vector Machines (SVM). The optimizations are performed using genetic algorithms and have been represented with the Research Object Optimization Ontology. An RDF representation of the Research Object can be downloaded from this link. An alternative representation in RDF-a can be found in this document.
Optimization Research Object: Aggregated Resources
The "Parameter Optimization of an Ecological Niche Modeling Workflow" Research Object aggregates several resources used to create the optimizations of the workflow. This section introduces each of the resources step by step and explains them in detail.
- The ENM workflow being optimized. It has been encoded and executed in Taverna, and it is currently uploaded in myExperiment. The workflow has several input parameters, and some of them have fixed values in all the optimization execution runs. These parameters are: Nu (value=0.5) , Coef0 (value=0.5) , Degree (value=3) , Kernel_Type (value=2) , SVM_Type (value=0) , ProbabilisticOutput (value=1) and Mask (value=wcs>http://biovel.iais.fraunhofer.de/geoserver/ows?>biovel_temp:SJB_Baltic_NorthEastAtlantic _20130522_112604_531) . The parameters being optimized are numberOfPseudoAbstences, Gamma and Cost.
- The Search Space Configuration, with the subworkflow on top of which the search is being performed: the "Select algorithm" subworkflow. The search space configuration for the parameters being optimized is as follows: Gamma has a minimum value 0, a maximum value 10 and round value true. Cost has a minimum value 0, a maximum value 8 and and exponential function with value (base) 2. Finally, numberOfPseudoAbstences has minimum value 200, and maximum value 600.
- A termination condition with maximum time 1440 minutes.
- The Genetic Algorithm used has two output parameters: Cross Over=0.75 and Mutation Rate=0.2 .
- The Fitness function used (single objective), which is performed on the "AUC" parameter.
- A selection of the optimization runs was performed on the Workflow, from more than 100 optimization runs. These runs are specified on the table below (note that they are not in chronological order of execution):
Optimization Run Number Cost Value Gamma Value NumberOfPseudoAbstences Value Fitness Value Run 1 8 2.36 363 0.9207 Run 2 64 5.87 458 0.9014 Run 3 8 6.84 319 0.8974 Run 4 32 3.79 486 0.9019 Run 5 256 8.94 568 0.9032 Run 6 32 0.06 215 0.9084 Run 7 8 4.78 491 0.8867 Run 8 8 1.25 562 0.9102 Run 9 64 6.09 514 0.8929 Run 10 4 2.24 453 0.9101
References
- [Guinan et al 2009]: J. Guinan, C. Brown, M. F. Dolan, and A. J. Grehan. Ecological niche modelling of the distribution of cold-water coral habitat using underwater remote sensing data. Ecological Informatics, 4(2):83-92, 2009.
- [Hutchinson 1957]: G. E. Hutchinson. Cold Spring Harbor Symposium on Quantitative Biology. Concluding remarks, 22:415-427, 1957.
- [Kulhanek et al 2011]: S. A. Kulhanek, B. Leung, and A. Ricciardi. Using ecological niche models to predict the abundance and impact of invasive species: application to the common carp. Ecological Applications, 21(1):203-213, 2011.
- [Wiley et al 2003]: E. O. Wiley, K. M. McNyset, A. T. Peterson, C. R. Robins, and A. M. Stewart. Niche modeling and geographic range predictions in the marine environment using a machine-learning algorithm. Oceanography, 16(3):120-127, 2003.
Authors of the Research Object
 |
Sonja Holl is a PhD student at the Juelich Supercomputing Centre (JSC) Forschungszentrum Juelich. She is the creator of the contents of the Research Object, and performed the optimizations on the parameters of the workflow. |
 |
Daniel Garijo is a PhD student in the Ontology Engineering Group at the Artificial Intelligence Department of the Computer Science Faculty of Universidad Politécnica de Madrid. He has contributed to the OWL encoding of the RO-Opt Ontology, extending the original Research Object Model and translating the conceptual Research Object Optimization to the model. He has also encoded this Research Object in html and RDF. |
 |
Khalid Belhajjame is a researcher at the University of Manchester. He has contributed to the OWL encoding of the RO-Opt Ontology, extending the original Research Object Model and the reviewing of this Research Object. |
Acknowledgements
The authors would like to give special thanks to Olav Zimmermann, Renato De Giovanni, Matthias Obst and Carole Goble for their help with the Research Object. The authors would also like to thank Alan Williams from the BioVEL project for his support on the DR work ow (BioVEL: EU FP7 283359 BioVel BioDiversity eLaboratory). This work was partly supported by the myGrid Platform Grant (EPSRC EP/G026238/1, "myGrid: A platform for e-Biology Renewal"), by the Wf4Ever European project (FP7-270192), and an FPU grant (Formacion de Profesorado Universitario) from the Spanish Science and Innovation Ministry(MICINN).

This page is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 2.0 Generic License.

